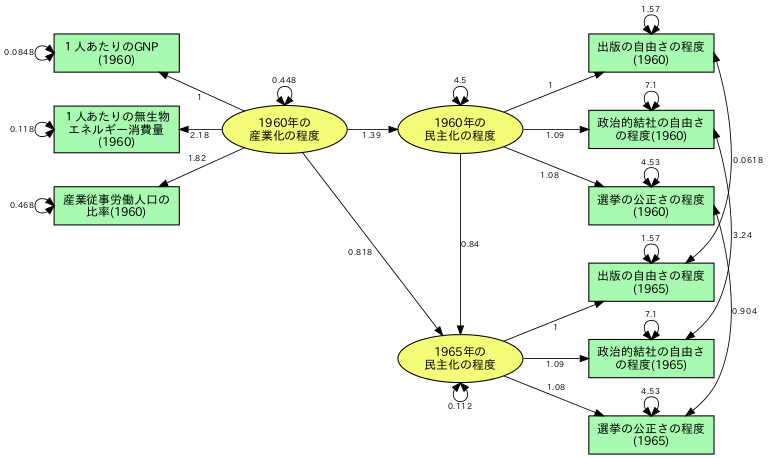
library(lavaan) mat<-' 0.54 0.99 2.28 0.82 1.81 1.98 0.73 1.27 0.91 6.89 0.62 1.49 1.17 6.25 15.58 0.79 1.55 1.04 5.84 5.84 10.76 1.08 2.06 1.58 5.06 5.60 4.94 6.83 0.85 1.81 1.57 5.75 9.39 4.73 4.987 11.38 0.94 2.00 1.63 5.81 7.54 7.01 5.82 6.75 10.80 ' wheaton.cov <- getCov(mat,names=c("x1","x2","x3","x4","x5","x6","x7","x8","x9")) model<-' F1=~x1+x2+x3 F2=~x4+a*x5+b*x6 F3=~x7+a*x8+b*x9 F2~F1 F3~F1+F2 x4~~c*x4 x7~~c*x7 x4~~x7 x5~~d*x5 x8~~d*x8 x5~~x8 x6~~e*x6 x9~~e*x9 x6~~x9 ' fit <- lavaan:::sem(model, sample.cov=wheaton.cov, sample.nobs=75) summary(fit, standardized=TRUE) latent<-c("F1","F2","F3") makeawk(latent) pars <- parameterEstimates(fit) g<-data.frame(pars$lhs,pars$op,pars$rhs," [label=",signif(pars$est,digits=3),"];") write.table(g,"out",sep=" ",col.names=FALSE,row.names=FALSE,quote=FALSE, na="") system( 'gawk -f "lavaan_dot.awk" "out">out.dot')
|